NIH Gives NC State ‘Outstanding Investigator’ a Grant to Advance Her Research on Cell Division
Dr. Caroline Laplante will use the R35 grant, more than $2 million over five years, to study the mechanisms that drive individual cells to divide into two. A better understanding of this cytokinesis process at its most fundamental level could lead to cancer breakthroughs down the line.

Dr. Caroline Laplante, an associate professor of quantitative and computational biology at the NC State College of Veterinary Medicine, has received an Outstanding Investigator Award from the National Institutes of Health to advance her work on unraveling the forces that drive cellular division.
The grant, called an R35 and funded through the NIH National Institute of General Medical Sciences, will provide Laplante and her lab more than $2 million over five years and is renewable. The R35, unlike an R01 NIH grant that is awarded to fund a specific research project, supports an individual investigator with an outstanding record of conducting research that holds promise.
Laplante’s research uses fission yeast to study how protein molecules inside a cell organize themselves into a ring that contracts and how this organization drives the separation of a mother cell into two daughter cells.
Little is known about how the contractile forces generated by this ring are transmitted to the plasma membrane, but Laplante’s earlier research has shown how protein clusters called nodes anchor the underlying actin filaments — which play a role in all cell shape changes including muscle contractions — to the plasma membrane.
These protein anchors play a crucial role in pulling the plasma membrane inward and causing the cell to separate. Ultimately, her research could lead to a better understanding of how organisms develop and cancers progress, processes that depend on both normal and aberrant cytokinesis.
“After the fertilization of an egg the cell undergoes its first division, and it’s going to divide into two, and then into four,” says Laplante, who has a B.Sc. in biochemistry and a Ph.D. in developmental biology. “That’s the normal process. When it becomes dysregulated, that’s when it leads to cancer and fibrotic diseases. You get masses and clumps. By studying the process at the very fundamental levels, you can understand the normal and then the dysregulated phases.”
Uncovering the forces that cause that first cell to divide is Laplante’s goal.
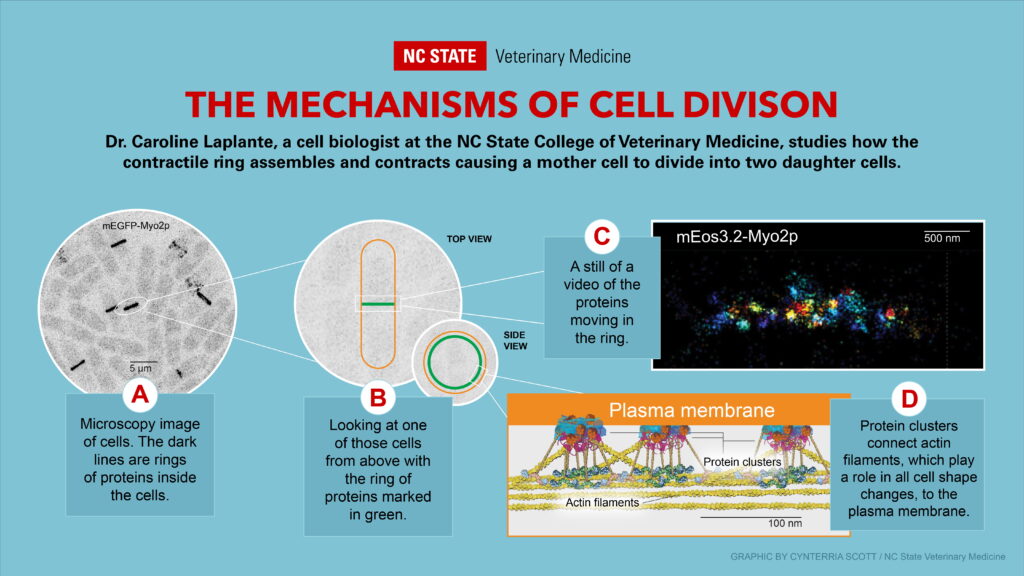
“That belt of proteins, if it’s not connected to the membrane of the cell, you’re just going to squeeze in on the inside of the cell and you’re not going to do anything,” she says. “So not only do you need these forces to act, but you need something to transmit these forces so that the cell changes shape.”
Laplante arrived as a faculty member at NC State University and the College of Veterinary Medicine’s Department of Molecular Biomedical Sciences in 2017 as part of a chancellor’s cluster of faculty hires in quantitative and computational developmental biology.
“NC State, at the college and at the campus level, does a really great job of recognizing talent, and supporting our faculty members to scientific independence is one of our key investments,” says Dr. Joshua Stern, associate director of research for the veterinary college. “With folks like Caroline, it’s paid off big time. She’s doing amazing work, and she’ll change the field based on her work.”
A TRAIL-BLAZER IN MICROSCOPY
As a postdoc, Laplante used what was then a new type of microscopy to discover that those protein complexes are the basic unit of constriction. The microscopy, now called Single Molecule Localization Microscopy (SMLM), used fluorescence to visualize subcellular structures at the nanoscale and led to a Nobel Prize in chemistry for its developers in 2014. SMLM had been used only on fixed samples when Laplante was one of the first to use the method in live cells.
With SMLM of live cells, Laplante can record time-lapse of cell division, showing for the first time at unprecedented resolution how proteins change and rearrange their organization. She and her team are building an analysis pipeline, named NanoTrax, to understand the dynamics of protein clusters in dense structures at the nanoscale.

“Now we can see their little motions as the cell divides and ask, ‘How is this protein complex behaving?’ And what are those behaviors revealing about their environments?” she says. “Answering these questions can tell us how these proteins work and reveal their role in the mechanisms that drive cytokinesis.”
Laplante and her team have already presented NanoTrax at international conferences, and she anticipates that her next published paper will detail how NanoTrax can visualize and measure the environments the protein clusters are in.
To understand how NanoTrax works, she provides an analogy of concertgoers in a crowd, trying to move around and continuously bumping into people.
“If you were to take a picture from above, and I have a light on my head, and you try to track me, you would see my motions are affected by the others around me,” she says. “That’s exactly what we’re doing. We’re labeling these protein clusters, and we’re measuring their motions in crowded environments.”
NanoTrax can determine whether protein clusters move directionally, undergo diffusive motions, undergo stop-and-go motions or change direction.
“Those directional versus nondirectional motions is what we’re after,” she says. “And down the road, NanoTrax can be used to answer many other biological questions in a variety of other contexts.”
Laplante continues to be one of the few scientists using SMLM in live cells because of the advanced level of mathematical and computational training required to extract usable information from the visuals that SMLM provides.
“This type of microscopy could be applied to many other things, but the problem is that big hurdle in math,” she says. “More and more of these microscopes are available commercially, but they are hard to use well without that extra level of training I got as a post-doc.”
Eventually, Laplante wants to study the protein movements of more complex organisms, possibly C. elegans, a tiny worm that lives in temperate soil and animal cells. The proteins that make the cell ring constrict are evolutionarily conserved, from fission yeast to animals, so the core mechanisms are also expected to be conserved, she says.
A FUNDAMENTAL QUESTION
Now that she has opened the hood on cell division, she says, she wants to see what happens when certain parts are missing.
“There are many pieces in the contractile ring, and if you take one out, what happens to the others?” she says. “We are just starting to remove pieces right now.”
Sometimes people are surprised that colleges of veterinary medicine include departments and researchers focusing on molecular biomedical sciences, but how cells divide is a fundamental question for all animals.

“It affects the development of all animals, and cancer also affects all animals,” Laplante says. “So if you’re looking at the very core mechanism of development, the answers are going to be shared, whether you’re trying to answer questions for humans or for any species.”
The NC State College of Veterinary Medicine is committed not only to providing outstanding clinical care, but also to uncovering the causes of diseases, says Dr. Michele Battle, head of the Department of Molecular Biological Sciences.
“That is why our department is home to a team of outstanding researchers, such as Dr. Laplante, with a mission to catalyze life-changing research,” Battle says. “Their discoveries about fundamental biological processes directly inform and accelerate the development of new preventions, diagnostics and treatments that benefit our animal patients and human health. They explain the why and how of disease and are crucial to future veterinary and human medical advances.”